JOURNAL OF CLINICAL GENETICS AND HEREDITY
On this page
Translational Pharmacogenomics Augmenting Pharmacotherapy
Kalyan Ram Uppaluri*, Hima Jyothi Challa, Vamsi Challa, Kavya Reddy Kothapally, Krishna Vardhani K, Natya K, Aswini K, Kalyani Palasamudram, K Sri Manjari and Anusha G
GenepoweRx, Suit #2B, Plot No. 240, Nirvana, Road No. 36, Jawahar Colony, Jubilee Hills, Telangana, Hyderabad, India
*Corresponding Author: Kalyan Ram Uppaluri, GenepoweRx, Suit #2B, Plot No. 240, Nirvana, Road No. 36, Jawahar Colony, Jubilee Hills, Telangana, Hyderabad, India.
| ReceivedJan 5, 2023 | RevisedJan 10, 2023 | AcceptedJan 15, 2023 | PublishedJan 23, 2023 |
Abstract
Pharmacogenomics (PGx), the intertwined knowledge of pharmacology and genomics, is well known. Taking PGx in pace with modern sequencing technological advancements is essential for effective clinical utilization, implementation, and personalized treatment care. Molecular profiling is a technique used to analyze the genetic and molecular makeup of an individual's cancer or other diseases. Decisions on a patient's course of treatment, including the choice of an appropriate pharmacotherapy, can be made using this information.
For example, molecular analysis can pinpoint genetic changes that increase a patient's propensity to respond to a particular treatment. It can also reveal potential resistance mechanisms, which helps choose effective treatments for patients. In addition, molecular profiling can help to identify possible side effects that a patient may experience when taking certain medications. Overall, molecular profiling can be a valuable tool in helping to optimize pharmacotherapy for an individual patient.
This review highlights the history of PGx, the role of pharmacogenes, PGx guidelines, the interrelation between PGx and personalized medicine, PGx technologies, challenges, and solutions in the implementation of PGx, international and national execution of PGx, and PGx in geriatric patient care, pediatric patient care, maternal, fetal medicine, general medicine, and oncology with a focus on the international and national scenarios. This review aims to better elucidate PGx for clinical implementation by healthcare workers, clinicians, and researchers to ensure efficient, safe, and cost-effective pharmacotherapeutic treatment plans for patients.
Keywords
Pharmacogenomics; Personalized medicine; PGx implementation; Molecular medicine; Targeted therapeutics; NGS; Molecular medicine; Targeted therapy
Introduction
Pharmacogenomics (PGx), a contemporary field of modern medicine is attaining the limelight at a quick pace as it paves the way for personalized medicine to become a reality. It combines knowledge from pharmacology and genomics and connects the patient’s genomic information, i.e., variations in an individual’s genetic code, with their response to specific drugs. This approach is beneficial to the clinician-patient duo. Clinicians can select the most appropriate drug for every individual with the best efficacy, correct dosage, and the slightest chance of potential side effects by gene-drug pairing for various medications, thus improving the treatment precision [1]. For the patients, it may be a cost-saving as well as a risk-mitigating strategy.
History of PGx
The recognition of PGx goes way back to 510 BC when Pythagoras noticed that ingesting fava beans caused hemolytic anemia and oxidative stress in only a few, but not all, individuals [2]. Only in the 1950s, when the field was more established, scientists understood the association between favism and linked to glucose-6-phosphate dehydrogenase deficiency [3]. Some of the discoveries made during the early periods of PGx include recognizing a variant of the butyrylcholinesterase enzyme, which causes prolonged paralysis after the use of succinylcholine, and a variant form of glucose-6-phosphate dehydrogenase deficiency (rs1050828) which can lead to hemolytic anemia after taking primaquine, an antimalarial drug [4].
Vogel coined the term PGx in 1959 [5]. Twin studies done in the 1960s comparing the genetic similarities in identical twins to fraternal twins supported the claim that genetics was involved in drug metabolism [6]. Concerning PGx in oncology, polymorphic responses to the anti-leukemic drug 6-mercaptopurine (6-MP) were identified by Weinshilboum and Sladek in 1980 [7]. Well-recognized examples of pharmacogenetic variation are the polymorphism of thiopurine S-methyltransferase (TPMT; *2 and *3 alleles) and CYP2C19 haplotypes (*2, *3, *17 alleles). Many predicted that the Human Genome Project’s launch in 2003 would speed up precision medicine by enhancing trial-and-error diagnosis and pharmaceutical treatment [8]. The first FDA-approved PGx test was AmpliChip™ CYP450, based on the Affymetrix Gene Chip platform using the microarray technology for assessment of an antiplatelet drug- clopidogrel’s metabolism and response which allowed the detection of 33 alleles in CYP2D6 and three alleles in CYP2C19 gene [9,10].
Role of pharmacogenes
Drugs administered orally or intravenously enter the bloodstream and reach the cells to perform their action. For the medication to function to its best potential, pharmacogenes should be free of functional mutations. These pharmacogenes encode proteins involved in the drugs ADME, i.e., absorption, distribution, metabolism, and excretion. Understanding the variations in these ADME genes serve as the basis of PGx [11]. Currently, the most common strategy in pharmacotherapy is to use the “one size fits all” approach for dosing and prescriptions, irrespective of the genetic differences in the drug response of individuals [12]. On average, approximately 40% of the medications taken by individuals every day are ineffective. A rough estimation is that the ineffectiveness of some medicines is over 50% [13]. To ensure a more appropriate treatment plan, understanding the principles of pharmacokinetics (PK) (i.e., ADME), pharmacodynamics (PD) (pharmacologic effects), and their relation to drug response, and then applying this knowledge in a clinical setting has been the significant lead for personalized medicine [1].
PGx guidelines
With leaps of innovation seen in next-generation sequencing technologies, PGx testing is becoming more and more accessible and affordable. Many PGx guidelines have been published by the international scientific consortia recently, such as the Clinical Pharmacogenetics Implementation Consortium (CPIC), Dutch Pharmacogenetics Working Group (DPWG), French National Network (Réseau) of Pharmacogenetics (RNPGx) and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) [14]. These are endorsed by various professional societies like ASHP-the American Society of Health-System Pharmacists; AMP-the Association for Molecular Pathology; and ASCPT-the American Society for Clinical Pharmacology and Therapeutics (ASCPT), and others. These guidelines have helped clinicians understand the genetic test result and tailor the pharmacotherapy leading to a better comprehension of the correlation between human health and genetic variation [15].
PGx and personalized medicine
Integration and implementation of PGx and personalized medicine are required to tailor pharmacological therapy and enhance its safety and effectiveness. Many individuals carry genetic variations that may lead to unwanted responses to specific drugs, putting them at risk of possible life-threatening events. For instance, a particular enzyme, having over 100 genetic variants with a continuum of activity, can metabolize codeine to morphine. Such children who may be ultrarapid metabolizers may get codeine-induced toxicity, which could be fatal. Physicians should consider PGX-guided medication to prevent occurrences of this nature [11,16]. Even in surgical settings, PGx-guided perioperative care not only informs anesthesiologists and surgeons on how to avoid adverse drug reactions (ADRs) caused by certain medications in patients but may also assist in pain management, thereby increasing patient satisfaction and reducing costs [17].
Technologies for PGx
Rapid and continuous progress in developing novel genotyping technologies over the last few decades, keeping in mind the possible advantages and limitations regarding PGx utility. The most commonly used methods, Sanger sequencing, and qPCR, can only detect targeted single nucleotide variants (SNV) or targeted exons as they contain a pre-selected panel of genetic variants, offering a short turnaround time, low costs and straightforward interpretation and are limited by its ability to identify rare variants. There are substantial disadvantages to Sanger sequencing in PGx, which screens many variations or locations for specific drug responses. These drawbacks include test optimization and turnaround times, which can be lengthy and impact test employment if therapeutic decisions are based on these results. A thermocycler, RT-PCR, or qPCR is generally employed to amplify sequences using SNV panels. Either commercially made or customized arrays may be utilized [18].
The other technology is a target enrichment method based on microarrays in which an oligonucleotide array directly hybridizes with DNA. Some limitations of this array-based approach are its ability to focus on relatively common variants, its inability to read variants with low minor allele frequencies (MAF), its need for hardware redesign every time a new gene variant gets added, and its lack of proper validation.
The confirmation of the selected variants ranges from small arrays with only specific variants of interest to massive arrays with all the potential pharmacogenetic variants. Most of the accessible arrays utilize PCR, sequencing by synthesis (SBS), beads, or nanospheres, in combination with chemiluminescence or fluorescence for the detection and identification of the kind of variant located at the site of interest [18]. Mass spectrometry could help to compare the difference in mass between a mutant and wild-type nucleotide [19]. Other technologies like next-generation sequencing (NGS) and long-read sequencing are favorable in PGx research as they generate data and allow the evaluation of haplotypes and structural variants compared to SNV panels. NGS may provide a colossal step towards personalized medicine, especially in cancer care, as it enables early somatic mutation detection, germline mutation detection, detection of resistance mechanisms, and mutational burden quantification, making it a cost- and time-effective treatment plan [20]. It also provides options for parallel and paired-end sequencing, which improves the diagnostic process and allows for optimal treatment strategy and therapeutic decision-making. A unique benefit to NGS is its ability to discover rare variants and novel variants and yield accurate quantitative results at a higher throughput scale. However, all the sequencing data cannot be made use of in clinical practice as of yet. The ultimate goal is to match the right technology to the suitable problem [18].
Challenges in integration and implementation of PGx
The significant prerequisites and challenges faced by the scientific and medical fraternity for integration of PGx into clinical testing are the cost factor, integral technology limitations, extensive population testing with data integration, and drafting regulatory frameworks for various technologies used in PGx [21,22]. Currently, as the cost of the test outweighs its benefits, clinics don’t seem to be keen on adopting this strategy [8]. Moreover, the implementation of PGx testing requires an accredited lab with scientific expertise to decode genomic information and integrate it with clinical data. Smaller clinics may be disadvantaged, particularly in rural areas lacking proper infrastructure. Though some companies are trying to develop PGx panels, commercialize them and implement PGx in clinics, due to their varying degrees of coverage, pharmacogenetics selected for molecular analysis, and algorithms used for interpreting results, the cost remains high. It is not affordable for every section of the community [23]. The variation in testing may attribute to a lack of validation, decreased standards of interpretation, and minimal regulatory independent oversight. Based on the analytical validation or genotyping accuracy, till now, only a single combinatorial commercial PGx test has been published [24]. Along with access to an equipped lab, selecting gene variants for patient sample testing is another challenge, as most genes have pleiotropic effects. Core ethical issues, including privacy and confidentiality, must also be addressed before practicing PGx [8]. Additionally, since the evidence and results from a genetic test may be suggestive, data access and patient autonomy are also areas of concern [25].
New allele variations, which are still being discovered and reported, require relevant validation for their use in clinical practice. Clinicians need to be well-versed in the evidence-supported utility of specific genes to make as accurate decisions as possible for choosing treatment plans [8]. Rare genetic variants may receive less research [25]. Hence, figuring out the rare alleles in specific populations is much needed to incorporate them into the testing panels, which means community-specific panels are more effective and accurate than the panels designed for global use. On the other hand, Haplotypes have posed a new challenge in PGx molecular testing. The metabolic phenotypes include normal, intermediate, poor, or rapid metabolizers. Studying the combined effect of these haplotypes and/or diplotypes is much more effective than evaluating the genetic status of an SNV associated with drug response or metabolism. However, there is a variety of evidence for the different gene pairs used in phenotype prediction; hence, allocating a metabolizer status is based on the probability of prevalence of a particular haplotype or haplotype subtype [26]. Proper infrastructure to reposit and report test results routinely, educating physicians on the benefits of testing, and getting third-party payments are other hurdles to be challenged to make PGx a personalized treatment option [16].
Solutions
Most drugs have a wide variety of pharmacokinetics enzymes that metabolize them and erratic action targets. Hence, combinatorial algorithms written by combining pharmacokinetic and pharmacodynamic gene variant’s genotype status, ultimately recommending one medication, will aid in having a comprehensive approach to drug response assessment. This is different from a multigene panel, where medicines are recommended based on the effects of the individual gene variant, one at a time [27]. Educating physicians, patients, nurses, and pharmacists are mandatory to improve healthcare quality. They must be well-versed in understanding genetic information and implementing it in treatment plans. Even though PGx testing could reduce the financial burden on the patient in the longer run, it is a preventive measure and, thus, may not be common knowledge to everyone. Therefore, the healthcare provider should discuss the purpose, benefits, limitations, and alternatives of genetic tests with the patient. Nurses and pharmacists should also be kept up to date on the possibilities and limitations of PGx to ensure high-quality patient care, especially since nurses have firsthand interactions with patients [8]. There is a necessity for strong institutional support, definite goals, and standardization of strategies and procedures to enlighten healthcare providers and patients. Such amalgamation will ultimately assist in driving PGx implementation to the clinical setting and help precision medicine thrive [28]. To overcome the knowledge gap over the last twenty years, pharmacy schools are implementing PGx-related courses that overlap in pharmacy and precision medicine. As 50% of the Clinical Pharmacogenetics Implementation Consortium implementers are led by pharmacies, these professionals are vital to integrating precision medicine into clinical practice [29]. Cumulative knowledge from pharmacists, molecular scientists, and clinicians makes this data more reliable and can bridge the gap between molecular technologies, molecular diagnosis, and clinical application to make personalized medicine a reality.
For the implementation of PGx in personalized medicine, there is also a requirement to construct an appropriate and secure IT infrastructure integrated with clinical decision support (CDS) in addition to more PGx evidence; strict regulations; strategies for reimbursing stakeholder’s acceptances; incorporating education in all clinics and institutes and promoting PGx to patients and healthcare providers [30]. CDS provides clinicians with information to utilize PGx to optimize pharmacotherapy systematically [31]. Integrating CDS into electronic health records (EHR) is beneficial as it increases PGx clinical knowledge and improves result interpretation. The first step is the identification of PGx biomarkers associated with pharmacological action, drug targets, drug metabolization, enzymes, and transporters. The next step is to characterize candidate genes in terms of their functional and non-functional variants, as well as their allele frequencies and ethnic variations. Following this step comes the design of algorithms, the selection of the gene-drug pair(s), the development of CDS, laboratory testing, and the verification of previously stated results. Before being used on independent populations, the response phenotypes, which may include metabolism, effectiveness, or toxicity, must be confirmed through a controlled study first. Then the therapeutic medicine, along with the associated diagnostic test, will be put on the market after receiving regulatory approval and completing confirmatory clinical trials. Distribute the therapeutic drug in conjunction with the diagnostic test companion. Engage with and notify the relevant stakeholders (clinicians, institute heads, lab specialists, bioinformaticians, PGx test interpreters, etc.), get facilities (PGx testing kits, EHR), and receive funds. Determine the clinical impact, then analyze the Pharmacoeconomics [32,33] (Figure 1).
PGx implementation-international scenario
A 2018 international survey, upgraded from a 2010 survey [35] consisting of 204 respondents from 43 countries (compared to the 53 responses from 28 countries in 2010), studied the challenges of clinical implementation of PGx in different parts of the world. Even with widespread availability and positive change in attitude towards PGx testing, there was no significant change in the challenges of clinical implementation, with cost, reimbursement, physician skepticism, insufficient evidence, vague guidelines, and interpretation of results being the main challenges.
The advent of new data management difficulties in North America impedes PGx performance in countries with variable Human Development Index.
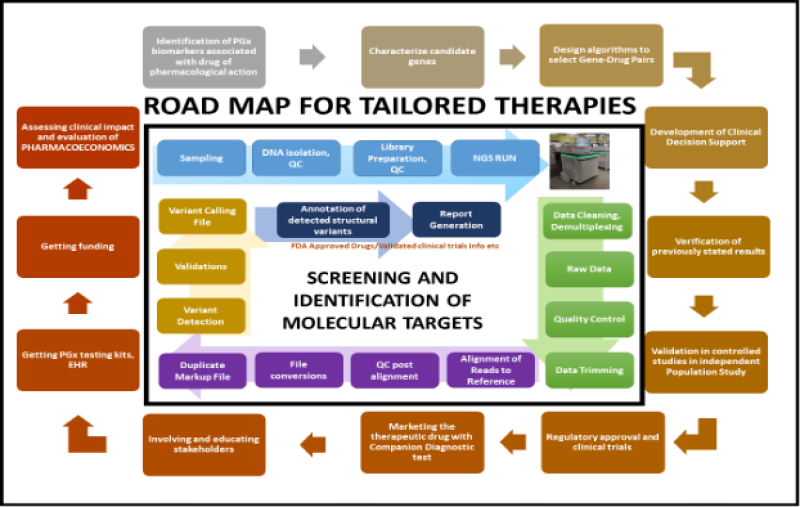
Figure 1: Steps to implement pharmacogenomics testing in clinical practice and implementing it in a population successfully.
These challenges arise due to the emergence of new problems in North America regarding CDS integration into HER [35]. CDS integration into EHR has a lag in test turnaround time, absence of standardization for genotyping tests and results, large-scale testing difficulties, test unavailability’s, human resources or infrastructure, low funding, and inadequate regulations. In a UK study, though the participants recognized advantages in the implementation of PGx, they also noted some limitations, such as integrating PGx with clinical practice and educating healthcare workers about the actual use and cost of PGx, which would benefit patients in the longer run. The social, legal, and ethical aspects and their impact on patients and the healthcare system should also be given importance to successfully implement it at a national and international level [36]. In the first-ever study for Flanders, done to assess the knowledge and applications of future physicians and pharmacists towards PGx in clinical practice, among 201 participants, the mean percentage of correct answers given on an assessment test (theoretical and practical) was only 34%. The small study suggests adequate education and knowledge exchange are required to introduce PGx into clinical practice [37].
PGx is currently not implemented in clinical practice in the West Bank of Palestine (WBP). When 381 physicians from various cities of WBP filled in a questionnaire, 81.1% responded that PGx was not an integral part of their medical education, showing that exposure to PGx was low. Only 58.5% of respondents agreed that PGx testing had relevance in their clinical practices. Also, >60% of respondents claimed they could not prescribe pharmacotherapy based on a PGx test. Thus, there is a need for PGx education and clinical implementation in WBP, especially the inclusion of PGx education in medical curriculums [38]. In sub-Saharan African (SSA) countries, clinical implementation of PGx testing can vastly improve healthcare quality, considering the high prevalence of communicable diseases, the increasing prevalence of non-communicable diseases, and the genetically diverse populations residing in SSA. The barriers to PGx clinical implementation include a lack of clinical care resources, a shortage of PGx clinical trials, social and ethical issues, and pharmacogenetic variant genotyping barriers. By investing in SSA. PGx research on a large scale, initiating biobanks/databases, and vitalizing PGx education to healthcare providers, PGx-guided treatment could become a reality in SSA [39]. A popular biotechnology company, 23andMe, based in California, USA, has put sufficient efforts into providing a direct-to-consumer online genomics testing service that uses SNP genotyping to test for genetic predispositions, traits, and even ancestry, after receiving a patient’s saliva sample. The tests are performed in CLIA-certified and CAP-accredited labs, and the kits are FDA-approved [40].
PGx implementation-national scenario
The first human genome sequencing in India, at the Institute of Genomics and Integrative Biology (IGIB), New Delhi, was a part of the Human Genome (HUGO) project. The Indian Genome Variation (IGV) was a CSIR-led project in which IGIB screened 1,000 genes having biomedical and pharmacogenetic importance in the genetic spectrum of India [41]. IGIB further completed complete genome screening of 1000 individuals pan India and highlighted the various PGx markers and their similarities and differences among the population of India. In recent years, Uppaluri Kandh Personalized Medicine Clinic, Hyderabad, India, has conducted PGx testing to provide a complete and comprehensive healthcare package for its patients. They combine genomic medicine with the latest advances in modern medicine to develop a personalized plan for each patient. GenepoweRx pharmacogenomics test is a PGx test done to tailor medications to the patient’s genetic makeup, enabling safe and effective treatments by determining ideal doses and minimizing side effects. This is mainly beneficial to three types of people,
1. patients with a chronic disease, such as diabetes, hypertension, heart conditions, gastrointestinal disorders, hormonal imbalances (thyroid disorders and fibromyalgia), inflammatory joint diseases, cognitive decline, premature aging, and chronic fatigue.
2. patients with early signs of chronic diseases, such as borderline blood sugar levels, energy loss, and obesity among others, and
3. patients looking to minimize side effects from their medications by learning about the right medications at the right doses for them.
Thus, pharmacogenomics test helps prevent the hassle of trial-and-error medications and educates the patients about the correct medication that suits their genetic profile based on their side effect status and likely side effects, metabolism profile, and dosage recommendations of specific medications and thus prevents treatment failure and ADRs. High-throughput genotyping and deeper sequencing coverage provides data accuracy. Med Genome in Bangalore and Genes2Me Pvt. Ltd. in Delhi, India, also offer similar services in India.
On the other hand, from the PGx education aspect, a study highlighted that out of the 138 MBBS students who participated in the cross-sectional study assessment from a medical college in Raipur, India, 95% of the students defined PGx correctly. However, only 54% knew the genetic variations in drug targets, transporters, and metabolizing enzymes that affect pharmacotherapy. As low as 15% knew about PGx testing availability in India, and 84% thought PGx education is a must and should be incorporated in the curriculum to understand screening technologies and biomarkers to focus on, interpretation of results, and application of PGx in clinical practice [12]. The JIPMER Integrated Pharmacogenomics Program (JIPP), held in Puducherry, India, conducted an Indo-Swiss symposium on advanced PGx strategies and personalized medicine implementation. The conference enabled Indian and Swiss researchers to interact, learn and bridge the gap between basic knowledge and recent advances in PGx [42].
PGx in general medicine
The treatment of several acute to chronic ailments such as asthma, diabetes, hypertension, kidney conditions, liver issues, and neurological problems, among others, could be done with the help of PGx guidance. Currently, the treatment regimens for type 2 diabetes mellitus (T2DM) include metformin as the first line of treatment, sulfonylureas, thiazolidinedione (TZD), and others as the second line of treatment. Even with this diversity of drug alternatives, there remains inter-patient variation due to variances in PGx [43]. This variability in drug response observed in T2DM patients is due to genetic variations of the genes which encode cytochrome P-450 enzymes and ATP-sensitive potassium channel proteins in the pancreatic beta cells. Studies have noted that the drug metformin is not suitable for all patients. On the other hand, in patients treated with metformin, variations in PK and PD metformin responses may be associated with genetic variations of the genes which encode organic metformin transporters. Genes encoding Solute Carrier Family proteins such as SLC47A1, SLC47A2, SLC2A2, SLC22A1, SLC22A2, SLC22A3 and SP1, FMO5, PPARA, AMHR2, CPA6, CAPN10, PRPF31 genes are studied to understand metformin metabolism, response, and toxicity. Specific drug targets and gene variants in patients with T2DM and obesity, such as GPCR genes, may be targeted for PGx studies to understand the nature of altered receptor function, thereby figuring out the compounds which will re-establish regular receptor function and proper drug responsiveness, which is a PGx-based personalized therapy for patients [44].
Even though there are various anti-hypertensive medicines on the market, only one-third of people diagnosed with hypertension have their blood pressure under control. It is necessary to conduct PGx research on anti-hypertensive drugs such as diuretics, calcium channel blockers, β-blockers, angiotensin-converting enzyme inhibitors, and angiotensin II receptor antagonists to guarantee individualized treatment [45]. Physicians should have information on how polymorphisms from different genes affect drug response pathways [46]. With PGx in lung-related issues, a randomized clinical trial (RCT) was conducted for 604 asthmatic patients, comparing nedocromil/budesonide versus placebo. PGx analysis inferred that distinctive effects with the placebo and drug treatments could guide the personalized development of drugs for asthmatic patients [47]. Four genes that show potential in PGx-implemented asthma therapy include FCER2, ABCC1, LTC4S, and ADRB2 [48]. Isoniazid PGx-guided therapy was shown to enhance health outcomes and save costs when treating drug-susceptible tuberculosis in Brazil, South Africa, and India. It was also a better option to standardize weight-based treatments [49]. The new global pandemic, coronavirus disease 2019 (COVID-19), has no direct PGx data available from patients. However, there are comprehensible mechanisms where genetic determinants could alter the outcome of currently used repurposed drugs for COVID-19. More PGx studies are required for these repurposed drug therapies to establish evidence-based guidelines for genetic testing [50]. Psychiatric disorders, such as depression, bipolar disorder, attention-deficit/hyperactivity disorder (ADHD), schizophrenia, narcolepsy, and epilepsy, may have PGx biomarkers like CYP2C19 and HLA-A [51]. Cacabelos et al. (2020) reported improper prescription of more than 50% of psychotropic drugs, highlighting the role of PGx in general medical practices [52].
PGx and geriatric patient care
PGx could assist clinicians in optimizing geriatric patient care while reducing adverse drug reaction (ADR) risks and simplifying complicated medication treatments. The implementation of PGx in geriatrics is a multilayered approach, including redesigning curriculums and providing more experiential and patient education [53]. Pharmacotherapy in elders includes polypharmacy, complex medicines, and handling inappropriate prescriptions that put older adults at a higher risk than younger adults. After a certain age, it is common knowledge that many of the body’s functions begin to deteriorate gradually. Though it is debatable whether genomic variations play a trial or a significant role in elders compared to younger individuals, intriguingly, for genomic variations in somatic cells in discernibly benign tissues, there is a possible noteworthy age- and disease-related increase. However, this acquired somatic mutation’s effect on drug efficacy or ADRs is still in the early stages of research [54]. In elders, many drug elimination pathways could be blocked or reduced by drug-drug interactions or due to the age factor [54], which could lead to ADRs. These ADRs and unwanted treatment outcomes may cause multimorbidity and geriatric syndromes. Approximately 25% of older adult’s adverse drug events were estimated to be preventable in ambulatory settings [53]. With regards to cardiovascular diseases (CVD) in elderly patients, beneficial PGx findings for most cardiovascular drugs have been reported, which suggest that preventive testing can enhance efficacy and reduce toxicity risk by recommending proper dosing. In the guidelines provided by the PGx consortium, one can find instructions on the correct administration of oral antiplatelet medicines, anticoagulants, beta-blockers, and statins. The need to produce tools for predicting multiple drug-drug-gene interactions is necessary as it’s prevalent in elderly comorbid patients, and currently, most only assess single drug-gene interactions [55]. Combinatorial PGx testing has proved to help select medications for older patients suffering from depression compared to the treatment-as-usual (TAU) approach [56]. Identifying actionable biomarkers can significantly improve elderly patient health [54].
PGx and pediatric patient care
Research on PGx in the pediatric population raises numerous unique ethical concerns; due to the invasive procedures required in blood sample collection, some parents may be hesitant to give their children their agreement to participate in the study. Replacing these with non-invasive saliva sample collection kits could also be used, and although the quality is the same for both types of samples, saliva is limited because it has reduced DNA quantity compared to blood [57]. The fact that most of the PGx data utilized in the generation of therapeutic guidelines by the Food and Drug Administration (FDA) and the National Institute of Health (NIH) concentrate on adult patients is another disadvantage of this method. Thus, extrapolating this data may be inaccurate in pediatric populations, and pediatric-specific research is needed [58].The DNA sequence remains constant throughout an individual’s lifetime. However, the expression of genes is not. Potentially they have a higher expression in early life stages than adulthood or vice versa. An example of such drug-metabolizing enzymes is the UDP glucanosyltransferase (UGT). UGT has only 1% of adult expression levels at birth and promptly increases to adult levels by 14 weeks [57].
A pharmacogenomics study investigated the advantages of whole-genome sequencing–guided testing over point-of-care drug–recommended testing in pediatric patients. In this 2-part-cohort study comprising 172 pediatric patients (mean age of 8.5 years), 36.8% were incompatible with the standard treatment process for the point-of-care cohort. For the whole-genome sequencing pre-emptive cohort, 80%) were advised of nonstandard treatment processes based on their pharmacogenetic profile [59].In pediatric oncology, PGx and personalized treatment plans are paramount to identify dosage requirements based on their genetic profile for the most favorable outcome and better quality of life for pediatric cancer survivors. As we know, chemotherapeutics has a narrow therapeutic window. PGx is used to determine the most appropriate course of action in pediatric oncology, which is necessary to personalize treatment plans for each patient because the severity of toxicity experienced by each patient varies depending on the individual [60]. For pediatric patients with kidney diseases, accurate and early diagnoses provide the best possible treatment options, as pediatric nephrology is a less acute specialty with a stronger focus on preventative care and counseling [61]. The short- and long-term benefits of PGx testing in children, especially with increasing evidence and a decrease in costs for multiple gene testing technologies, should influence a positive attitude toward uptake and user testing. However, for clinical utility, more pediatric trials need to be conducted to understand the impact of drug response in children, who at that young age still do not have a stable expression for several genes related to drug metabolism and transport [62].
PGx and maternal-fetal care
Due to the physiological and metabolic changes that occur during gestation, which genetic factors may further implicate, PGx may be helpful for prescribing medication for pregnant women. Research on the drug disposition by PGx and pharmacokinetic influences is still insufficient. On the other hand, it is common knowledge that several enzymes involved in metabolism have different activity levels during pregnancy. Consequently, a gynecologist can take the assistance of PGx during gestation for medication prescription [63]. A few biomarkers that are PGx liabilities during pregnancy include G6PD, CYP2C19, CYP2D6, and HLA, among others [62,63]. The use of thiopurine drugs, such as azathioprine and 6-mercaptopurine, by the mother for the treatment of inflammatory bowel disease (IBD) may provide a little danger to the fetus; however, PGx test can evaluate the amount of risk [64]. A well-timed aspirin dosage during pregnancy can significantly cut the probability of developing preeclampsia. Utilizing PGx allows for individualized therapy, dosage, and administration timings, which are all critical for reducing the risk of maternal and fetal morbidity and mortality [65].
PGx in oncology
A tumor is a hub for genetic mutations; hence, even though all the cancers known to date are the same, every tumor is genetically diverse. Understanding the genome signature of tumor cells provides authentic information to target cancer cells with exceptional precision. Targeted therapy makes this possible by using drugs specific to a few molecular targets to identify and attack tumorigenic cells more precisely. With the advent of FDA approval in 1970 [66] for tamoxifen in breast cancer treatment, a new era began in understanding the heterogeneity of tumor cell microenvironments in various cancers, which paved the way for personalized therapies in cancer treatment. Signal transduction inhibitors, Hormone therapies- in the case of breast and prostate cancer, angiogenesis inhibitors to prevent the spread of cancer, immunotherapy with monoclonal antibodies, and gene expression modulators are a few targeted therapies approved by FDA for cancer treatment.
The Memorial Sloan Kettering Cancer Institute and the MD Anderson Cancer Center in the United States are examples of institutes that conduct extensive research on target medicines and put these therapies into practice. The NDK project (National Decade against Cancer), which is a collaborative initiative between the German Federal Ministry of Education and Research (BMBF) and other stakeholders operating in the field of cancer research, is also working towards the creation and advancement of treatments for cancer patients in the hopes of improving their quality of life and increasing their chances of survival. Germany is now known to have the maximum use of targeted therapies for PD-1 and PD-L inhibitors after the USA, with more than two-fold increased use than the UK [67]. The French government, too, has given a high priority to the national cancer control program and to optimizing cancer care. As a part of this, it raised innovation funds for the RIHN (Referentiel des actes innovants hors nomenclatures) to improve molecular diagnosis and implement targeted therapies. Four molecular signatures, namely, Oncotype DX (21 gene-signature panels), Prosigna/PAM 50 (50 gene-signature panels), Endopredict (12 gene-signature panels), and Mammaprint (70 gene-signature panels) had obtained conditional access as of 2018. The American Joint Committee on Cancers (AJCC), the National Comprehensive Cancer Network (NCCN), the American Society of Clinical Oncology (ASCO), and the European Society of Medical Oncology are responsible for the development of PGx guidelines for cancer therapy (ESMO). GENEpower OncoRx Companion Diagnostics by Uppaluri KandH personalized medicine clinic is another effort to bridge the gap in translational medicine in India. It identifies molecular targets and recommends tailored therapies for patients with cancer. The examined gene targets play a crucial role in the disease’s pathophysiology and contribute to patient stratification. It is a directed test for drug decision-making licensed therapeutic solutions and aids in selecting the appropriate drug to prevent treatment failure and adverse effects. It also aids in forecasting the response and resistance to medicine and the selection and management of immunotherapy. This NGS-based in-vitro diagnostic genomic test targets DNA and RNA variants from Formalin-Fixed Paraffin-Embedded (FFPE) tumor tissue samples. A comprehensive footprint of many genes, including the regulatory regions, SNPs of entire coding regions, CNVs, and selective rearrangements such as translocations, genomic signatures with microsatellite instability (MSI), and tumor mutational burden (TMB), among others, which are candidate cancer biomarkers and potential onco-therapeutics with proven evidence from various clinical trials. Figure 1 depicts the brief workflow for screening and identifying molecular targets for targeted therapies. Personalized medicine in cancer patient care plays a significant role in cancer patients’ prevention, prognosis, and diagnosis. It is a preventative approach and forestalls disease progression [68]. Cancer chemotherapy also dramatically benefits from PGx as it provides preliminary molecular diagnostics for individualized chemotherapeutic care, which increases efficacy and reduces toxic side effects [69]. However, unlike PGx in general medicine, PGx in oncology helps identify the molecular target for therapies. Tailored drug’s risks and side effects are hard to discuss, even though they guarantee a better treatment strategy.
Conclusion
Most healthcare professionals do not apply PGx testing in their clinical practices. With more technological advances, education, and awareness of the benefits of PGx testing, its implementation in clinical settings should favorably increase. This review shows the effectiveness of PGx testing in all community sectors, from geriatric to pediatric patient care. There are still several unanswered questions regarding PGx testing and its implementation, one significant doubt being its incremental value for widespread use. It is necessary to discuss and resolve the ethical issue of whether PGx testing may lead to stigmatization, exclusion, or discrimination in particular communities.
It is essential to look into the unavailability of safe drug options for some genetic profiles. Given the right conditions in possible implementation settings, PGx could play a crucial role in many facets of medicine. This review focuses on generic issues with potential solutions applicable everywhere.
References
1. Saunders H, Harris D, Chirilă RM. Pharmacogenomics: Introduction and Use in Clinical Practice. Rom J Intern Med. 2020;58(2):69-74. PubMed | CrossRef
2. Pirmohamed M. Pharmacogenetics and Pharmacogenomics. Br J Clin Pharmacol. 2001;52(4):345. PubMed | CrossRef
3. Prasad K. Role of Regulatory Agencies in Translating Pharmacogenetics to the Clinics. Cases Miner Bone Metab. 2009;6(1):29.
4. Johnson JA. Pharmacogenetics: Potential for Individualized Drug Therapy Through Genetics. Trends Genet. 2003;19(11):660-6. PubMed |CrossRef
5. Vogel F. Moderne Probleme Der Humangenetik. In Ergebnisse Der Inneren Medizin and Kinderheilkunde. 1959;52-125.
6. Motulsky AG, Qi M. Pharmacogenetics, Pharmacogenomics and Ecogenetics. J Zhejiang Univ Sci B. 2006;7(2):169-70. PubMed | CrossRef
7. Weinshilboum RM, Sladek SL. Mercaptopurine Pharmacogenetics: Monogenic Inheritance of Erythrocyte Thiopurine Methyltransferase Activity. Am J Hum Genet. 1980;32(5):651.
8. Liu JC, Gorbovskaya I, Bousman C, Brown LC, Müller DJ. Opportunities and Challenges of Implementation Models of Pharmacogenomics in Clinical Practice. Per Med. 2020:449-57.
9. Squassina A, Artac M, Manolopoulos VG, Karkabouna S, Lappa-Manakou C, Mitropoulos K, et al. Translation of Genetic Knowledge into Clinical Practice: The Expectations and Realities of Pharmacogenomics and Personalized Medicine. Pharmacogenomics. 2010;11(8):1149-67. PubMed | CrossRef
10. Giusti B, Saracini C, Galora S, Marcucci R. Pharmacogenomics of Clopidogrel (Chapter 25). Handbook Of Pharmacogenomics and Stratified Medicine. AP. 2014;509-541.
11. Orrico KB. Basic Concepts in Genetics and Pharmacogenomics for Pharmacists. Drug Target Insights. 2019;1177392819886875. PubMed | CrossRef
12. Agrawal M, Kirtania L, Jha A, Hishikar R. Students’ Knowledge and Views on Pharmacogenomic Education in the Medical Curriculum. Indian J Pharmacol. 2021;53(1):19. PubMed | CrossRef
13. Banerjee M. Is Pharmacogenomics A Reality? Challenges and Opportunities for India. Indian J Hum Genet. 2011;17(Suppl 1):S1. PubMed | CrossRef
14. Abdullah-Koolmees H, Van Keulen AM, Nijenhuis M, Deneer VH. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and Rnpgx Guidelines. Front Pharmacol. 2021;11:595219. PubMed | CrossRef
15. Cecchin E, Stocco G. Pharmacogenomics and Personalized Medicine. Genes (Basel). 2020;11(6):679. PubMed | CrossRef
16. MOC C. Pharmacogenomics: An Evolving Clinical Tool for Precision Medicine. Cleve Clin J Med. 2020;87(2):91. PubMed | CrossRef
17. Kaye AD, Koress CM, Novitch MB, Jung JW, Urits I, Viswanath O, et al. Pharmacogenomics, Concepts for the Future of Perioperative Medicine and Pain Management: A Review. Best Pract Res Clin Anaesthesiol. 2020;34(3):651-62. PubMed | CrossRef
18. Van der Lee M, Kriek M, Guchelaar HJ, Swen JJ. Technologies for Pharmacogenomics: A Review. Genes (Basel). 2020;11(12):1456. PubMed | CrossRef
19. Gabriel S, Ziaugra L, Tabbaa D. SNP Genotyping Using the Sequenom Massarray Iplex Platform. Curr protoc Hum Genet. 2009;60(1):2-12. PubMed | CrossRef
20. Ruiz-Garcia E, Astudillo-de la Vega H, editors. Translational Research and Onco-Omics Applications in the Era of Cancer Personal Genomics. Adv Exp Med Biol. 2019; 1168:9-30 PubMed | CrossRef
21. Hippman C, Nislow C. Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges. J Pers Med. 2019;9(3):40. PubMed | CrossRef
22. Verbelen M, Weale ME, Lewis CM. Cost-Effectiveness of Pharmacogenetic-Guided Treatment: Are We There Yet? the pharmacogenomics J. 2017;17(5):395-402. PubMed | CrossRef
23. Bousman CA, Hopwood M. Commercial Pharmacogenetic-Based Decision-Support Tools in Psychiatry. Lancet Psychiatry. 2016;3(6):585-90. PubMed | CrossRef
24. Jablonski MR, King N, Wang Y, Winner JG, Watterson LR, Gunselman S et al. Analytical Validation of a Psychiatric Pharmacogenomic Test. Per Med. 2018;15(03):189-97. PubMed | CrossRef
25. Corrigan OP. Pharmacogenetics, Ethical Issues: Review of the Nuffield Council on Bioethics Report. J Med Ethics. 2005;31(3):144-8. PubMed | CrossRef
26. Eum S, Lee AM, Bishop JR. Pharmacogenetic Tests for Antipsychotic Medications: Clinical Implications and Considerations. Dialogues Clin Neurosci. 2016;18(3):323-337 PubMed | CrossRef
27. Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical Validity: Combinatorial Pharmacogenomics Predicts Antidepressant Responses and Healthcare Utilizations Better than Single Gene Phenotypes. Pharmacogenomics J. 2015;15(5):443-51. PubMed | CrossRef
28. Wang YT, Merl MY, Yang J, Zhu ZX, Li GH. Opportunities for Pharmacists to Integrate Pharmacogenomics into Clinical Practice. Pharmacogenomics J. 2020;20(2):169-78. PubMed | CrossRef
29. Nagy M, Eirini Tsermpini E, Siamoglou S, Patrinos GP. Evaluating the Current Level of Pharmacists’ Pharmacogenomics Knowledge and Its Impact on Pharmacogenomics Implementation. Pharmacogenomics. 2020;21(16):1179-89 PubMed | CrossRef
30. Klein ME, Parvez MM, Shin JG. Clinical Implementation of Pharmacogenomics for Personalized Precision Medicine: Barriers and Solutions. J Pharm Sci. 2017;106(9):2368-79. PubMed | CrossRef
31. Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating Pharmacogenomics into Electronic Health Records with Clinical Decision Support. Am J Health Syst Pharm. 2016;73(23):1967-76. PubMed | CrossRef
32. Lam YW. Scientific Challenges and Implementation Barriers to Translation of Pharmacogenomics in Clinical Practice. ISRN Pharmacol. 2013;2013 PubMed | CrossRef
33. Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin Transl Sci. 2016;9(5):233. PubMed | CrossRef
34. Ghaddar F, Cascorbi I, Zgheib NK. Clinical Implementation of Pharmacogenetics: A Nonrepresentative Explorative Survey to Participants of World pharma 2010. Pharmacogenomics. 2011;12(7):1051-9. PubMed | CrossRef
35. Abou Diwan E, Zeitoun RI, Abou Haidar L, Cascorbi I, Khoueiry Zgheib N. Implementation and Obstacles of Pharmacogenetics in Clinical Practice: An International Survey. Br J Clin Pharmacol. 2019;85(9):2076-88. PubMed | CrossRef
36. Rafi I, Crinson I, Dawes M, Rafi D, Pirmohamed M, Walter FM. The Implementation of Pharmacogenomics into UK General Practice: A Qualitative Study Exploring Barriers, Challenges and Opportunities. J Community Genet. 2020;11(3):269-77. PubMed | CrossRef
37. Edris A, Vanoverschelde A, Bushaj P, Van Nieuwerburgh F, Lahousse L. Pharmacogenetics in Clinical Practice: Current Level of Knowledge Among Flemish Physicians and Pharmacists. Pharmacogenomics J. 2021;21(1):78-84. PubMed | CrossRef
38. Jarrar Y, Musleh R, Hamdan A, Ghanim M. Evaluation of the Need for Pharmacogenomics Testing Among Physicians in the West Bank of Palestine. Drug Metab Pers Ther. 2021;36(4):289-94. PubMed | CrossRef
39. B. Tata E, A. Ambele M, S. Pepper M. Barriers to Implementing Clinical Pharmacogenetics Testing in Sub-Saharan Africa. A Critical Review. Pharmaceutics. 2020;12(9):809. PubMed | CrossRef
40. Lu M, Lewis CM, Traylor M. Pharmacogenetic Testing Through the Direct to Consumer Genetic Testing Company 23 and Me. BMC Med Genomics. 2017;10(1):1-8. PubMed | CrossRef
41. Ambedkar SR, Yadav P. Pharmacogenomics Current Scenario: Clinical Applications and Challenges.
42. Xaviar S, Kumar S, Ramasamy K, Chakradhara Rao US. Indo–Swiss Symposium on Advances in Pharmacogenomic Strategies for Implementation of Personalized Medicine. Pharmacogenomics. 2021;22(2):67-71. PubMed | CrossRef
43. Guo Z, Priefer R. Current Progress in Pharmacogenomics of Type 2 Diabetes: A Systemic Overview. Diabetes and Metabolic Syndrome. Diabetes Metab Syndr. 2021;15(5):102239. PubMed | CrossRef
44. Gao W, Jockers R. Pharmacogenomics of GPCR Genes in Type 2 Diabetes and Obesity. Curr Opin Endocr Metab Res. 2021;16:128-35.
45. Oliveira-Paula GH, Pereira SC, Tanus-Santos JE, Lacchini R. Pharmacogenomics and Hypertension. Pharmgenomics. Pers Med. 2019;12:341. PubMed | CrossRef
46. Luizon MR, Pereira DA, Sandrim VC. Pharmacogenomics of Hypertension and Preeclampsia: Focus on gene–gene interactions. Front Pharmacol. 2018;9:168. PubMed | CrossRef
47. Wang RS, Croteau‐Chonka DC, Silverman EK, Loscalzo J, Weiss ST, Hall KT. Pharmacogenomics and Placebo Response in a Randomized Clinical Trial in Asthma. Clin Pharmacol Ther. 2019;106(6):1261-7. PubMed | CrossRef
48. García-Menaya JM, Cordobés-Durán C, García-Martín E, Agúndez JA. Pharmacogenetic Factors Affecting Asthma Treatment Response Potential Implications for Drug Therapy. Front Pharmacol. 2019;10:520. PubMed | CrossRef
49. Rens NE, Uyl-de Groot CA, Goldhaber-Fiebert JD, Croda J, Andrews JR. Cost-Effectiveness of a Pharmacogenomic Test for Stratified Isoniazid Dosing in Treatment of Active Tuberculosis. Clin Infect Dis. 2020;71(12):3136-43. PubMed | CrossRef
50. Badary OA. Pharmacogenomics and COVID-19: Clinical Implications of Human Genome Interactions with Repurposed Drugs. Pharmacogenomics J. 2021;21(3):275-84. PubMed | CrossRef
51. Kam H, Jeong H. Pharmacogenomic Biomarkers and Their Applications in Psychiatry. Genes (Basel). 2020;11(12):1445. PubMed | CrossRef
52. Cacabelos R. Pharmacogenomics of Drugs Used to Treat Brain Disorders. Expert Rev Precis Med Drug Dev. 2020;5(3):181-234.
53. Roman YM, Dixon DL, Salgado TM, Price ET, Zimmerman KM, Sargent L, et al. Challenges in Pharmacotherapy for Older Adults: A Framework for Pharmacogenomics Implementation. Pharmacogenomics. 2020;21(9):627-35. PubMed | CrossRef
54. Brockmöller J, Stingl JC. Multimorbidity, Polypharmacy and Pharmacogenomics in Old Age. Pharmacogenomics. 2017;18(6):515-7. PubMed | CrossRef
55. Božina N, Vrkić Kirhmajer M, Šimičević L, Ganoci L, Mirošević Skvrce N, Klarica Domjanović I, et al. Use of Pharmacogenomics in Elderly Patients Treated for Cardiovascular Diseases. Croat Med J. 2020;61(3):147-58. PubMed | CrossRef
56. Forester BP, Parikh SV, Weisenbach S, Ajilore O, Vahia I, Rothschild AJ, et al. Combinatorial Pharmacogenomic Testing Improves Outcomes for Older Adults with Depression. Am J Geriatr Psychiatry. 2020;28(9):933-45. PubMed | CrossRef
57. Hawcutt DB, Thompson B, Smyth RL, Pirmohamed M. Paediatric Pharmacogenomics: An Overview. Arch Dis Child. 2013;98(3):232-7. PubMed | CrossRef
58. Parry CM, Hawcutt D. Fifteen-Minute Consultation: Pharmacogenomics: A Guide for Busy Clinicians. Arch Dis Child Educ Pract Ed. 2020;105(2):107-10. PubMed | CrossRef
59. Cohn I, Manshaei R, Liston E, Okello JB, Khan R, Curtis MR, et al. Assessment of the Implementation of Pharmacogenomic Testing in a Pediatric Tertiary Care Setting. JAMA Netw Open. 2021;4(5):e2110446. PubMed | CrossRef
60. Bernsen EC, Hagleitner MM, Kouwenberg TW, Hanff LM. Pharmacogenomics as a Tool to Limit Acute and Long-Term Adverse Effects of Chemotherapeutics: An Update in Pediatric Oncology. Front Pharmacol. 2020;11:1184. PubMed | CrossRef
61. Quinlan C. The Future of Paediatric Nephrology Genomics and Personalised Precision Medicine. Curr Pediatr Rep. 2020;8(3):115-21.
62. Haga SB. Pharmacogenomic Testing in Pediatrics: Navigating the Ethical, Social, and Legal Challenges. Pharmgenomics Pers Med. 2019;12:273. PubMed | CrossRef
63. Betcher HK, George Jr AL. Pharmacogenomics in Pregnancy. Semin Perinatol. 2020;44(3):151222. PubMed | CrossRef
64. Andoh A, Kawahara M, Imai T, Tatsumi G, Inatomi O, Kakuta Y. Thiopurine Pharmacogenomics and Pregnancy in Inflammatory Bowel Disease. J Gastroenterol. 2021;56(10):881-90. PubMed | CrossRef
65. Saravia-Sucaticona M, Martinez-Garcia FE, Solar-Cuba MJ. The Timing of Aspirin Administration in Pregnancy is Important to Prevent Preeclampsia. Am J Obstet Gynecol MFM. 2021;3(4):100313. PubMed | CrossRef
66. Jordan VC. Effect of Tamoxifen (ICI 46,474) On Initiation and Growth Of DMBA-Induced Rat Mammary Carcinomata. Eur J Cancer (1965). 1976;12(6):419-24. PubMed | CrossRef
67. Gill J, Fontrier AM, Miracolo A, Kanavos P. Access to Personalised Oncology in Europe. LSE. 2020.
68. Ravichandiran N, Barman MK, Lavuri ST, Srivastava M, Sakthivel S, Singh M, et al. Precision Medicine in Cancer. In Handbook of Research on Advancements in Cancer Therapeutics. 2021;433-66. IGI Global.
69. Cheok MH, Evans WE. Acute Lymphoblastic Leukaemia: A Model for the Pharmacogenomics of Cancer Therapy. Nat Rev Cancer. 2006;6(2):117-29.
Kalyan Ram Uppaluri*, Hima Jyothi Challa, Vamsi Challa, Kavya Reddy Kothapally, Krishna Vardhani K, Natya K, Aswini K, Kalyani Palasamudram, K Sri Manjari and Anusha G
GenepoweRx, Suit #2B, Plot No. 240, Nirvana, Road No. 36, Jawahar Colony, Jubilee Hills, Telangana, Hyderabad, India
*Corresponding Author: Kalyan Ram Uppaluri, GenepoweRx, Suit #2B, Plot No. 240, Nirvana, Road No. 36, Jawahar Colony, Jubilee Hills, Telangana, Hyderabad, India.
Copyright© 2023 by Uppaluri KR, et al. All rights reserved. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Uppaluri KR, Challa HJ, Challa V, Kothapally KR, Vardhani K, Natya K, et al. Translational Pharmacogenomics Augmenting Pharmacotherapy. J Clin Genet Hered. 2023;1(1):1-18. DOI: https://doi.org/10.37191/Mapsci-JCGH-1(1)-001
