JOURNAL OF ENDOCRINOLOGY AND METABOLISM RESEARCH
Association of Gut Microbiota with the Incidence of Metabolic Syndrome among Type 2 Diabetes Mellitus Patients
Vipin Porwal1*, Himanshu Jain1 and Arun Maseeh2
1Department of Medicine, RD Gardi Medical College, Ujjain, Madhya Pradesh, India
2MD, Internal Medicine, Ahmedabad, Gujarat, India
*Corresponding Author: Vipin Porwal, MD, Professor, Department of Medicine, RD Gardi Medical College, Ujjain, Madhya Pradesh, India.
| ReceivedApr 1, 2024 | RevisedApr 9, 2024 | AcceptedApr 16, 2024 | PublishedApr 25, 2024 |
Abstract
Background: Alteration of the gut microflora leads to local intestinal inflammation which produces various primary and secondary metabolites that cause chronic inflammation, leading to metabolic syndrome.
Objectives: To determine the gut microbiome about biochemical and inflammatory markers in type 2 diabetes patients and to assess the clinical outcomes of metabolic syndrome for gut microbiota composition.
Methods: In this hospital-based observational study, 41 type 2 diabetes mellitus patients were included and underwent clinical examination. The gut microbiome diversity was detected from stool samples through DNA extraction followed by 16s RNA gene sequencing and bioinformatical analyses. Using blood samples, the immune response analysis was also done by identifying CD4+T cells through blood immune cells analysis.
Results: After gene sequencing from stool samples, the gut microbiota species of rumonicoccus, fusobacterium, and blautia were identified among 9(42.9%), 9(42.9%), and 10(47.6%) patients only in the case group. However, fecalibacterium prasunitzii, bifidobacterium, akkermansia mucinphila and bacteriodes fragilis were 1(5%), 2(10%), 10(50%) patients only in the control group. In both groups, lactobacillus, and escherichia coli were seen among 3(15%) and 20(100%) patients in the control group, followed by 4(19%) and 20(95.2%) patients in the type 2 diabetes group. The incidence of metabolic syndrome was seen in 4(20%) patients in the control group and it was in 19(90.5%) patients in the case group where the prevalence of metabolic syndrome was significantly higher in the type 2 diabetes group than the control group (Fisher’s Exact Test, P=0.001).
Conclusion: Among the studied population, the gut microbiota was found to be significantly different in the diabetics when compared to non-diabetics and helped to identify the bacteria that were important in the development of diabetes. The elevation of inflammatory markers further showed the possible role of gut microbiota in creating a state of chronic inflammation in these patients, ultimately leading to insulin resistance and the development of type 2 diabetes mellitus.
Keywords
Probiotics; Insulin resistance; Chronic inflammation; Metabolic syndrome; Diabetes mellitus, Gut microbiota
Introduction
The diabetes and its global prevalence affects about 537 million people worldwide. In 2021, it was estimated that one in ten adult individuals was affected by this multifactorial disease [1]. These multiple factors include smoking, hypertension, sleep, dyslipidemia, family history, stress, and sedentary lifestyle [2]. In addition, gut microbiota has turned out to be an important contributor to the development of type 2 diabetes mellitus (T2DM) [3-6]. The gut microbiota has an essential function in several metabolic processes, including the digestion and absorption of undigested carbohydrates, minerals and electrolytes, the regulation of bowel motility, and the production of certain micro-nutrients [2]. The mechanism by which gut microbes influence glucose metabolism and insulin resistance is associated with insulin resistance, obesity and T2DM [7,8]. These inflammations have been linked to increased production of inflammatory cytokines such as Tumor necrosis factor-alpha (TNF-α), Interleukin-1 (IL-1) and IL-6 and nuclear factor kappa light chain enhancer of activated B cells (NF-kB) [8]. Studies have shown that B cell function was severely compromised by increased IL-1 while inflammatory biomarker C reactive protein is increased by translocation of IL-6 into the liver [9]. The reduction in inflammation was found when T2DM subjects were supplemented with probiotics specifically with lactobacillus, bifidobacterium clostridium, and akkermansia [10,11]. These subjects suggested supplementation of probiotics with specific microbes may serve as an important therapeutic application in the management of type 2 Diabetes. Further, over the past decade, significant efforts have been made to map the structural and functional attributes of human gut microbial communities to understand the disease progression [12,13]. Throughout life, the gut microbiota acts as a sensory hub, responding to both intrinsic and extrinsic stimuli affecting host physiology within and outside the gut [14]. Disruption of a delicate balance among the gut microbes has been linked to the development of metabolic diseases particularly type 2 diabetes mellitus [5], obesity [15], and cardiovascular disorders [16]. Most of the earlier studies reported differences between the gut microbiome of diabetics, prediabetics, and healthy non-diabetic individuals [6,17], and very few examined the gut microbiome of treatment-naive T2DM individuals [18,19]. This study aimed to identify the possible role of gut microbes in the development of diabetes mellitus, which will pave the way for path-breaking interventions in the management of diabetes.
Materials and methods
This was a hospital-based observational study carried out at R.D. Gardi Medical College and Charitable Hospital, Ujjain in the Department of General Medicine, over 2 years. The sample size was determined by using this formula, n=Z2*P*(100-P)/d2 where Z=1.96 at 95% confidence interval, P=36% (Prevalence of diabetes mellitus, 36%) [13] and absolute error, d=15%. Thus, the minimum sample size determined was 40 cases. Patients of age above 18 yrs and below 80 yrs who came to the outpatient department or those who were admitted to the hospital diagnosed with a case of T2DM were included in the study whereas, those who have chronic kidney disease, coronary artery disease, congestive cardiac failure, urinary tract infection, stroke, ketonuria, overt diabetic nephropathy, pregnant and non-cooperative patients were excluded. A pre-structured proforma was used to collect baseline data and a detailed history with clinical examination was also done. Further, the stool samples were meticulously collected and transported through a reverse cold chain which was then subjected to DNA extraction followed by 16s RNA gene sequencing and bioinformatical analyses to detect gut microbiome diversity. Using blood samples, the immune response analysis was also done by identifying CD4+T cells through blood immune cells analysis.
The data were analyzed by using SPSS version 23.0. Statistical significance between the two groups was investigated by unpaired ‘t’ test. Continuous variables were expressed as mean, median, and range. Fischer’s exact test was employed to determine the influence of variables on the study variables. All tests were performed with P<0.05 considered statistically significant.
Results
About 41 patients were included where 20 (48.8%) patients in the control group and 21 (51.2%) patients in the T2DM group. The mean age (SD) was 52.55 (8.35) years in the control group and 52.43 (8.16) years in the case group. The mean fasting blood sugar (FBS) level in the control group was 99.40 ± 8.01 mg/dL and in the T2DM group, it was 187.19 ± 62.58 mg/dL where the difference was found to be statistically significant (P=0.001). The mean FBS was significantly higher in the T2DM group than in the control group. The mean glycated hemoglobin (HbA1c) in the control group was 5.05 ± 0.41% and in type 2 diabetes, it was 8.76 ± 2.27% of P=0.001 which was found to be statistically significant. Results showed that the mean HbA1c was significantly higher in the T2DM group than in the control group. The comparison of mean haematological parameters between control and case group showed that the mean (SD) value of Hb was 12.23 (1.49) gm%, neutrophils of 5441.70 (1906.74) cells/µL, lymphocytes of 2154.45 (1050.07)%, platelets of 269100(63841) cells/µL, erythrocyte sedimentation rate (ESR) of 15.95 (5.13)mm/hour, serum glutamic pyruvic transaminase (SGPT) of 36.65 (11.79) IU/L, serum creatinine of 0.91 (0.20) mg/dL in the control group, whereas, in case of a group, the mean (SD) value of Hb was 11.68 (1.98 ) gm%, neutrophils of 7710.48 (4643.53) cells/µL, lymphocytes of 3631.52 (2073.81)%, platelets of 25100 (93586.32) cells/µL, ESR of 35.19 (23.57) mm/hour, SGPT of 51.91 (108.33) IU/L, serum creatinine of 2.06 (2.99) mg/dL as shown in Table 1.
|
Parameters |
Control group [Mean ± SD] (n=20) |
Case group [Mean ± SD] (n=21) |
‘t’ value, df |
P value |
|
Hb (gm%) |
12.23 ± 1.49 |
11.68 ± 1.98 |
0.984, df=39 |
0.331 |
|
Neutrophils (cells/µL) |
5441.70 ± 1906.74 |
7710.48 ± 4643.53 |
-2.027, df=39 |
0.049* |
|
Lymphocyte (%) |
2154.45 ± 1050.07 |
3631.52 ± 2073.81 |
-2.855, df=39 |
0.007* |
|
Platelets (/µL) |
269100 ± 63841 |
251000 ± 93586.32 |
0.720, df=39 |
0.476 |
|
ESR (mm/hour) |
15.95 ± 5.13 |
35.19 ± 23.57 |
-3.568, df=39 |
0.001* |
|
SGPT (IU/L) |
36.65 ± 11.79 |
51.91 ± 108.33 |
-0.626, df=39 |
0.535 |
|
Serum creatinine (mg/dL) |
0.91 ± 0.20 |
2.06 ± 2.99 |
-1.720, df=39 |
0.093 |
Table 1: Comparison of mean hematological parameters between control and case group. ‘*’ represents the P value by unpaired ‘t’ test of P <0.05 was considered statistically significant. Hb: Haemoglobin; ESR: Erythrocyte Sedimentation Rate; SGPT: Serum Glutamic Pyruvic Transferase.
The mean hs CRP in the control group was 8.01 ± 4.62 mg/dL; in the T2DM group, it was 31.21 ± 44.86 mg/dL where their difference was statistically significant (P=0.027). Further, the result showed that mean hs CRP was significantly raised in the case group than the control group. The comparison of lipid profile between the control and T2DM groups showed that the mean (SD) values of total cholesterol, triglycerides, LDL cholesterol, VLDL cholesterol were higher with 199.33 (49.38) mg/dL, 193.35 (72.24) mg/dL, 123.25 (51.16) mg/dL, 40.41 (14.46) mg/dL in the case group than in the control group of 161.50 (17.2) mg/dL, 129.70 (25.73) mg/dL, 98.50 (16.90) mg/dL, 27.32 (5.27) mg/dL respectively. However, the mean HDL cholesterol was significantly reduced in the case group of 34.45 mg/dL than the control group of 50.33 mg/dL as depicted in Figure 1.
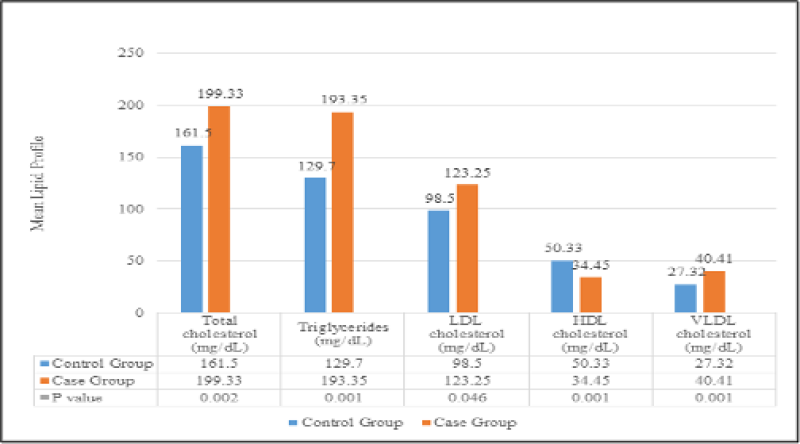
Figure 1: Comparison of mean lipid profile between the control and case group. P value by unpaired ‘t’ test of P <0.05 was considered statistically significant. LDL: Low-Density Lipoprotein; HDL: High-Density Lipoprotein; VLDL: Very Low-Density Lipoprotein.
After gene sequencing, the gut microbiota species was identified where rumonicoccus, fusobacterium and blautia were seen among 9 (42.9%), 9 (42.9%), 10 (47.6%) patients only in the case group which indicates that there was a higher prevalence of rumonicoccus, fusobacterium and blautia in T2DM patients. Contrarily, fecalibacterium prasunitzii, bifidobacterium, akkermansia mucinphila and bacteriodes fragilis were 1 (5%), 2 (10%), 10 (50%) patients only in the control group. Further, lactobacillus and escherichia coli were seen in both groups of 3 (15%) and 20 (100%) patients in the control group, followed by 4 (19%) and 20 (95.2%) patients in the T2DM group as shown in Table 2.
Gut microbiota species Group P value
|
Gut microbiota species |
Group |
P value |
|
|
Control Group (%) |
Case Group (%) |
||
|
Rumonicoccus |
0.00% |
42.90% |
0.001* |
|
Fecalibacterium prasunitzii |
5.00% |
0.00% |
0.488 |
|
Fusobacterium |
0.00% |
42.90% |
0.001* |
|
Bifidobacterium |
10.00% |
0.00% |
0.232 |
|
Akkermansia mucinphila |
35.00% |
0.00% |
0.003* |
|
Lactobacillus |
15.00% |
19.00% |
0.529 |
|
Bacteriodes Fragilis |
50.00% |
0.00% |
0.001* |
|
Blautia |
0.00% |
47.60% |
0.001* |
|
E. coli |
100.00% |
95.20% |
0.512 |
Table 2: Identification of gut microbiota species in control and case groups. ‘*’ represents the P value by Fisher’s Exact test where P<0.05 was taken as statistically significant.
The incidence of metabolic syndrome was seen in 4 (20%) patients in the control group and it was in 19 (90.5%) patients in the case group where the prevalence of metabolic syndrome was significantly higher in the T2DM group than control group (Fisher’s Exact Test, P=0.001).
Discussion
The current study was designed to analyze the gut microbiome about biochemical and inflammatory markers in type 2 diabetes patients and to evaluate the clinical outcomes of metabolic syndrome concerning the composition of gut microbiota. We found that gut microbiota species absent such as fecalibacterium prasunitzii, bifidobacterium, akkermansia mucinphila, and bacteriodes fragilis, and the presence of rumonicoccus, fusobacterium, and blautia only in type 2 diabetes patients. Along with that, there was a higher incidence of metabolic syndrome and lipid profile biomarkers among the type 2 diabetes group. It was noted that there was a raised level of mean hs CRP in type 2 diabetes patients as highlighted by Stanimirovic J et al. where numerous prospective studies reported the association between serum CRP level and risk of incident T2DM. Elevated CRP may serve as an unintentional risk factor for the progression of T2DM because it is unmistakably linked to the onset of prediabetes and diabetes-related vascular complications. Along with elevated FPG and HbA1C levels, abnormal OGTT results, hyperinsulinemia, and other factors, elevated CRP levels should also be taken into account when assessing the overall risk of T2DM [20].
The mean lipid profile biomarkers such as total cholesterol, triglycerides, LDL cholesterol, and VLDL cholesterol were elevated and the mean HDL was decreased in the case group which was correlated with Bhambhani GD as the frequencies of the high cholesterol, high TG, and high LDL levels were higher in the diabetic group, thus indicating that diabetic patients were more prone for dyslipidemia, which could cause cardiovascular disorders [21]. About the gut microbiota, rumonicoccus and fusobacterium were seen in none of the patients in the control group, and 9 (42.9%) type 2 diabetes patients of P=0.001. It was noted in a review by Gurung M et al. that pathobionts that may be harmful in T2DM, such as fusobacterium nucleatum and ruminococcus gnavus can elevate several inflammatory cytokines, albeit in different inflammatory disorders. According to the review, ruminococcus, fusobacterium, and blautia have a positive relationship with type 2 diabetes [6].
One (5%) patient in the control group and none of the T2DM patients had fecalibacterium prasunitzii. Fisher's exact test P value for the proportional difference was determined to be 0.488, indicating that it was statistically insignificant. In four out of five human case-control studies, it was discovered that F. prasunitzii was negatively associated with T2DM [22-26]. Bifidobacterium was seen in 2 (10%) patients in the control group and in none of the patients in the T2DM group. The proportional difference was found to be statistically not significant (Fisher’s exact test, P value=0.232). Bifidobacterium appears to be the genus most consistently supported by the literature as the one harbouring microorganisms potentially protective against T2DM, according to a review by Gurung M et al [6]. Moreover, akkermansia mucinphila was identified in 7 (35%) patients only in the control group of P value=0.003. The prevalence of akkermansia mucinphila was significantly higher in control group patients. In studies conducted by Zhang X [22] and Greer [27], the negative association between the abundance of this bacterium and T2DM has been reported in human studies.
Lactobacillus was noted in 3 (15%) patients in the control group and 4 (19%) of the patients in the T2DM group. The proportional difference was statistically not significant (Fisher’s exact test, P=0.529). The mean of these parameters when compared between the presence or absence of lactobacillus were found to be statistically not significant (P>0.05), which shows that the biochemical and inflammatory parameters are not affected by the presence or absence of lactobacillus. Gurung M claimed that when taking into account all association studies, including those that examined changes following treatments, the lactobacillus genus provides a complex situation of seemingly contradictory results [6].
However, in five out of six studies, cross-sectional investigations comparing patients and controls found a connection between the abundance of this species and T2DM [28-32]. Bacteroides fragilis was seen in 10 (50%) patients only in the control group with a statistically significant P value=0.001. The prevalence of bacteriodes fragilis was significantly higher in control group patients. Since over-expression of this cytokine in the muscle protects against age-related insulin resistance. Dagdeviren S et al. hypothesized that induction of IL-10 by species of roseburia intestinalis, bacteroides fragilis, akkermansia muciniphila, lactobacillus plantarum, and L. casei may improve glucose metabolism [33]. Blautia was seen only in 10 (47.6%) patients in the case group of P=0.001. The prevalence of blautia was significantly higher in T2DM patients. In three of the four cross-sectional investigations for T2DM, the Blautia genus was shown to be more prevalent in illness groups [22,34-36]. E. coli was seen in 20 (100%) patients in the control group, and in 20 (95.2%) patients in the T2DM. The proportional difference was statistically not significant (Fisher’s exact test, P=0.512).
In addition, lymphocytes were found to be significantly higher in a patient in whom E. coli was absent in comparison to the patients in whom E. coli was present (P=0.001), while the rest of the parameters when compared between the presence or absence of E. coli were found to be statistically not significant (P>0.05). E. coli presence had a significant effect only on lymphocytes, while the rest of the parameters were not affected by its presence.
The results of E. coli cannot be considered accurate, as there was only 1 patient in whom E. coli was absent which reduces the reliability of statistical analysis. In the control group, metabolic syndrome was seen in 4 (20%) patients and in the case group, it was seen in 19 (90.5%) patients. The proportional comparison was found to be statistically significant (Fisher’s Exact Test P=0.001). The prevalence of metabolic syndrome was significantly higher in the T2DM group than in the control group. In Type 2 Diabetes, metabolic syndrome was not present in 2 patients, while it was present in 19 patients. Even though there were large proportional differences between the presence and absence of metabolic syndrome in each organism, the comparisons did not reach statistical significance (P>0.05).
A study conducted by He Y provides evidence concerning the relationship between gut microbiota and metabolic diseases. This study discovered a positive correlation between metabolic disorders and fusobacterium, a significant colonic carcinogenic bacterium, suggesting that this bacterium may contribute to both metabolic disorders and colon cancer. After adjusting for the consumption of pre-, pro-, and synbiotics, lactobacillus was found to be positively associated with metabolic disorders, which is consistent with earlier reports that this bacterium is enriched in obese patients as well as in patients with type 2 diabetes, metabolic syndrome, stroke, and rheumatoid arthritis. Similarly, it has been discovered that bifidobacterium had a favorable correlation with FBG [37].
Strengths of this study
To the best of the author’s knowledge, this was the first study conducted by determining the gut microbiome of biochemical & inflammatory markers in type 2 diabetes patients and evaluating the outcomes of metabolic syndrome concerning the composition of gut microbiota. Further, it was confirmed that gut microbiota species such as rumonicoccus, fusobacterium, and blautia are present only in type 2 diabetes patients. In addition, there was an increased risk of metabolic syndrome and lipid profile biomarkers elevation among the type 2 diabetes group.
Limitations and future recommendations
Our study had a few limitations such as a smaller sample size compared to other similar studies and was conducted in a single center only. So, a multi-center study with a large sample size, of different ethnicity and dietary patterns might provide more specific results.
Conclusion
This study concluded that the gut microbiota was found to be significantly different in diabetics when compared to non-diabetics and helped to identify the bacteria that were crucial in the development of diabetes and differentiate them from the bacteria that played a protective role against diabetes. Moreover, the cases had significantly elevated inflammatory markers, and derangement of lipid profile was also found to be significant which further indicates the possible role of gut microbiota in the creation of a state of chronic inflammation in these patients that ultimately leads to insulin resistance and development of type 2 diabetes mellitus.
Ethical consideration
This study was approved by the Institutional Ethics Committee, R.D.Gardi Medical College, Ujjain (IEC-RDGMC) with reference number 24/2021.
Conflict of interest
Nothing to disclose.
Financial support
All authors have declared that no financial support was received from any organization for the submitted work.
References
1. Magliano DJ, Boyko EJ, Atlas ID. What is Diabetes? InIDF Diabetes ATLAS. 10th edition 2021. International Diabetes Federation.
2. Palau-Rodriguez M, Tulipani S, Isabel Queipo-Ortuño M, Urpi-Sarda M, Tinahones FJ, Andres-Lacueva C. Metabolomic Insights into the Intricate Gut Microbial–host Interaction in the Development of Obesity and Type 2 Diabetes. Front Microbiol. 2015;6:154996. PubMed | CrossRef
3. Wang X, Xu X, Xia Y. Further Analysis Reveals New Gut Microbiome Markers of Type 2 Diabetes Mellitus. Antonie Van Leeuwenhoek. 2017;110(3):445-53. PubMed | CrossRef
4. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut Bacteria Selectively Promoted by Dietary Fibers Alleviate Type 2 Diabetes. Science. 2018;359(6380):1151-6. PubMed | CrossRef
5. Toniolo A, Cassani G, Puggioni A, Rossi A, Colombo A, Onodera T, et al. The Diabetes Pandemic and Associated Infections: Suggestions for Clinical Microbiology. Rev Med Microbiol. 2019;30(1):1-7. PubMed | CrossRef
6. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine. 2020;51:102590. PubMed | CrossRef
7. Scheithauer TP, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, Van Raalte DH, et al. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front Immunol. 2020;11:571731. PubMed | CrossRef
8. Sikalidis AK, Maykish A. The Gut Microbiome and Type 2 Diabetes Mellitus: Discussing a Complex Relationship. Biomedicines. 2020;8(1):8. PubMed | CrossRef
9. Hajjo R, Sabbah DA, Al Bawab AQ. Unlocking the Potential of the Human Microbiome for Identifying Disease Diagnostic Biomarkers. Diagnostics (Basel). 2022;12(7):1742. PubMed | CrossRef
10. Sharifi-Rad J, Rodrigues CF, Stojanović-Radić Z, Dimitrijević M, Aleksić A, Neffe-Skocińska K, et al. Probiotics: Versatile Bioactive Components in Promoting Human Health. Medicina. 2020;56(9):433. PubMed | CrossRef
11. Salles BI, Cioffi D, Ferreira SR. Probiotics Supplementation and Insulin Resistance: A Systematic Review. Diabetol Metab Syndr. 2020;12:1-24. PubMed | CrossRef
12. Huda MN, Kim M, Bennett BJ. Modulating the Microbiota as a Therapeutic Intervention for Type 2 Diabetes. Front Endocrinol. 2021;12:632335. PubMed | CrossRef
13. Cunningham AL, Stephens JW, Harris DA. Intestinal Microbiota and their Metabolic Contribution to Type 2 Diabetes and Obesity. Gut Pathog. 2021;20(2):1855-70. PubMed | CrossRef
14. Zhang L, Chu J, Hao W, Zhang J, Li H, Yang C, et al. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediators Inflamm. 2021;2021:5110276. PubMed | CrossRef
15. Simeon GG, Ibemologi A, Cameron DG. Assessment of Possible Link of Intestinal Microbiota and Type 2 Diabetes Mellitus. Am J Mol Biol. 2021;11(3):63-72.
16. Das T, Jayasudha R, Chakravarthy S, Prashanthi GS, Bhargava A, Tyagi M, et al. Alterations in the Gut Bacterial Microbiome in People with Type 2 Diabetes Mellitus and Diabetic Retinopathy. Sci Rep. 2021;11(1):2738. PubMed | CrossRef
17. Maskarinec G, Raquinio P, Kristal BS, Setiawan VW, Wilkens LR, Franke AA, et al. The Gut Microbiome and Type 2 Diabetes Status in the Multiethnic Cohort. Plos One. 2021;16(6):e0250855. PubMed | CrossRef
18. Yang HT, Liu JK, Xiu WJ, et al. Gut Microbiome-Based Diagnostic Model to Predict Diabetes Mellitus. Bioengineered. 2021;12:12521-34. PubMed | CrossRef
19. Zhang S, Cai Y, Meng C, et al. The Role of the Microbiome in Diabetes Mellitus. Diabetes Res Clin Pract. 2021;172:108645. PubMed | CrossRef
20. Stanimirovic J, Radovanovic J, Banjac K, Obradovic M, Essack M, Zafirovic S, et al. Role of C-Reactive Protein in Diabetic Inflammation. Mediators Inflamm. 2022;2022:3706508. PubMed | CrossRef
21. Bhambhani G, Bhambhani RG, Thakor NC. Lipid Profile of Patients with Diabetes Mellitus: A Cross Sectional Study. Int J Res Med Sci. 2015;3(11):3292-95.
22. Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PloS One. 2013;8(8):e71108. PubMed | CrossRef
23. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut Metagenome in European Women with Normal, Impaired and Diabetic Glucose Control. Nature. 2013;498(7452):99-103. PubMed | CrossRef
24. Remely M, Aumueller E, Merold C, Dworzak S, Hippe B, Zanner J, et al. Effects of Short Chain Fatty Acid Producing Bacteria on Epigenetic Regulation of FFAR3 in Type 2 Diabetes and Obesity. Gene. 2014;537(1):85-92. PubMed | CrossRef
25. Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential Adaptation of Human Gut Microbiota to Bariatric Surgery–Induced Weight Loss: Links with Metabolic and Low-Grade Inflammation Markers. Diabetes. 2010;59(12):3049-57. PubMed | CrossRef
26. Graessler J, Qin Y, Zhong H. Metagenomic Sequencing of the Human Gut Microbiome Before and After Bariatric Surgery in Obese Patients with Type 2 Diabetes: Correlation with Inflammatory and Metabolic Parameters. Pharmacogenomics J. 2013;13:514-22. PubMed | CrossRef
27. Greer RL, Dong X, Moraes AC, Zielke RA, Fernandes GR, Peremyslova E, et al. Akkermansia Muciniphila Mediates Negative Effects of IFNγ on Glucose Metabolism. Nat Commun. 2016;7(1):13329. PubMed | CrossRef
28. Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, et al. Modulation of Gut Microbiota Dysbioses in Type 2 Diabetic Patients by Macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116(1):80-93. PubMed | CrossRef
29. Sedighi M, Razavi S, Navab-Moghadam F. Comparison of Gut Microbiota in Adult Patients with Type 2 Diabetes and Healthy Individuals. Microb Pathog. 2017;111:362-9. PubMed | CrossRef
30. Wu X, Ma C, Han L. Molecular Characterisation of the Faecal Microbiota in Patients with Type II Diabetes. Curr Microbiol. 2010; 61:69-78. PubMed | CrossRef
31. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling Type 2 Diabetes and Metformin Treatment Signatures in the Human Gut Microbiota. Nature. 2015;528(7581):262-6. PubMed | CrossRef
32. Ni Y, Mu C, He X, Zheng K, Guo H, Zhu W. Characteristics of Gut Microbiota and its Response to a Chinese Herbal Formula in Elder Patients with Metabolic Syndrome. Drug Discov Ther. 2018;12(3):161-9. PubMed | CrossRef
33. Dagdeviren S, Jung DY, Friedline RH, Noh HL, Kim JH, Patel PR, et al. IL-10 Prevents Aging-Associated Inflammation and Insulin Resistance in Skeletal Muscle. FASEB J. 2017;31(2):701-10. PubMed | CrossRef
34. Lippert K, Kedenko L, Antonielli L, Kedenko I, Gemeier C, Leitner M, et al. Gut Microbiota Dysbiosis Associated with Glucose Metabolism Disorders and the Metabolic Syndrome in Older Adults. Benef Microbes. 2017;8(4):545-56. PubMed | CrossRef
35. Egshatyan L, Kashtanova D, Popenko A, Tkacheva O, Tyakht A, Alexeev D, et al. Gut Microbiota and Diet in Patients with Different Glucose Tolerance. Endocr Connect. 2016;5(1):1-9. PubMed | CrossRef
36. Inoue R, Ohue-Kitano R, Tsukahara T, Tanaka M, Masuda S, Inoue T, et al. Prediction of Functional Profiles of Gut Microbiota from 16S rRNA Metagenomic Data Provides a More Robust Evaluation of Gut Dysbiosis Occurring in Japanese Type 2 Diabetic Patients. J Clin Biochem Nutr. 2017;61(3):217-21. PubMed | CrossRef
37. He Y, Wu W, Wu S, Zheng HM, Li P, Sheng HF, et al. Linking Gut Microbiota, Metabolic Syndrome and Economic Status Based on a Population-level Analysis. Microbiome. 2018;6:1. PubMed | CrossRef
Vipin Porwal1*, Himanshu Jain1 and Arun Maseeh2
1Department of Medicine, RD Gardi Medical College, Ujjain, Madhya Pradesh, India
2MD, Internal Medicine, Ahmedabad, Gujarat, India
*Corresponding Author: Vipin Porwal, MD, Professor, Department of Medicine, RD Gardi Medical College, Ujjain, Madhya Pradesh, India.
Copyright© 2024 by Porwal V, et al. All rights reserved. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Porwal V, Jain H, Maseeh A. Association of Gut Microbiota with the Incidence of Metabolic Syndrome among Type 2 Diabetes Mellitus Patients. J Endo Metabol Res. 2024;5(1):9-18.
DOI: https://doi.org/10.37191/Mapsci-2582-7960-5(1)-035
